Research
Genetic architecture of age-related chronic diseases
| Genetic predisposition is critical for complex human diseases. In my latest study, I performed a comprehensive mapping of genetic architecture of kidney disease by applying large scale GWAS, eQTL and meQTL analysis. I discovered that epigenome (DNA methylation) explains a larger fraction of heritability than gene expression. To further identify disease-causal genes, I proposed a multi-stage prioritization strategy and prioritized >500 kidney disease genes, including SLC47A1, whose causal role was defined in knockout mice model and in human individuals carrying loss-of-function variants. (Liu et al., 2022 Nature Genetics; Project page; Github). | 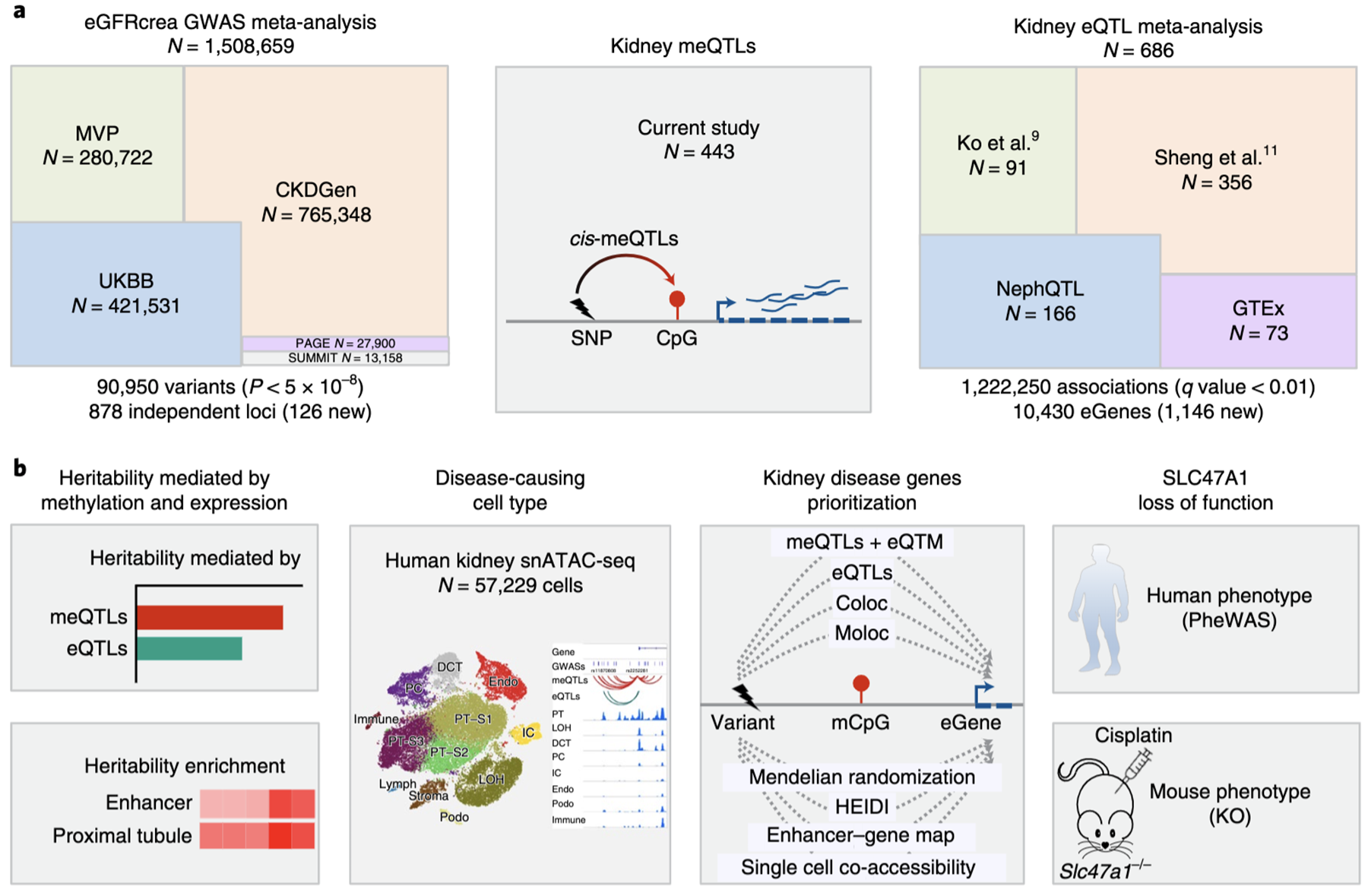 |
Cell type-specific epigenetic regulatory elements
| Epigenetic regulatory elements play important roles in cell fate determination. To identify these elements, I developed a series of bioinformatic software, such as SMART for de novo identifying tissue/cell-specific methylated regions from whole genome bisulfite sequencing data (Liu et al. 2016 NAR; Project page); QDMR for identifying differentially methylated regions from array-based methylation data (Zhang, Liu et al. 2011 NAR; Project page); QDCMR for identifying differential chromatin regions from ChIP-seq data (Liu et al. 2013 Sci. Rep.; GitHub). | 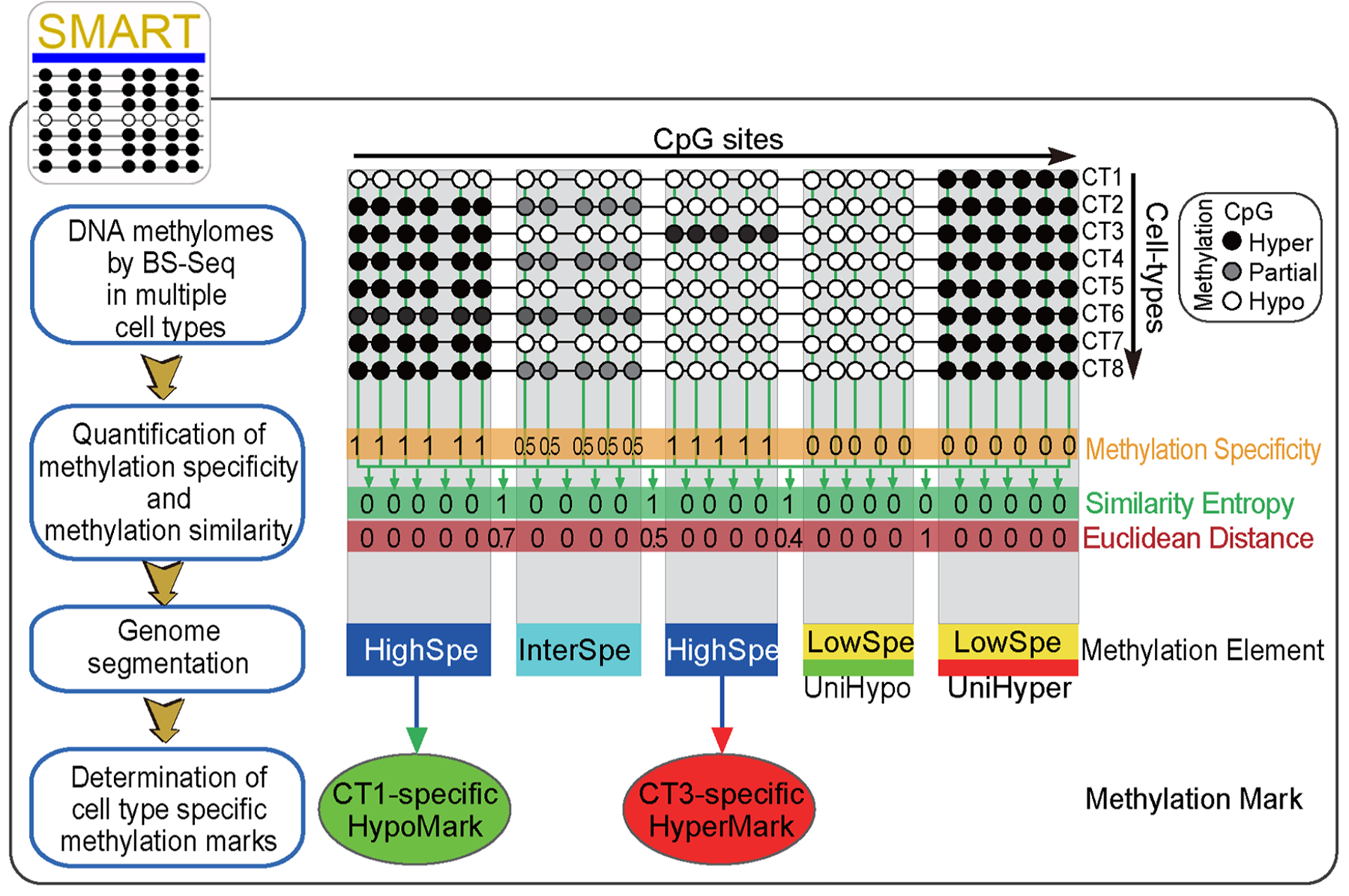 |
Epigenetic regulation of age-related chronic diseases
| Epigenetic regulators play central roles in complex diseases. By applying our bioinformatic tools to large-scale epigenome data, I identified disease-critical epigenetic regulatory elements, including DNA methylation, histone modifications, super-enhancers, and long non-coding RNAs, and explored their roles in complex human diseases (e.g. kidney disease, diabetes, and cancer) (Xu, Liu et al. 2019 Cell Death & Disease; Xiong,…, Liu et al. 2017 NAR; Wei,…, Liu et al. 2016 NAR; Lv, Liu et al. 2013 NAR; Lv, Liu et al. 2012 NAR; Zhang, Liu et al. 2011 NAR; Zhang, Lv, Liu et al. 2010 NAR). | 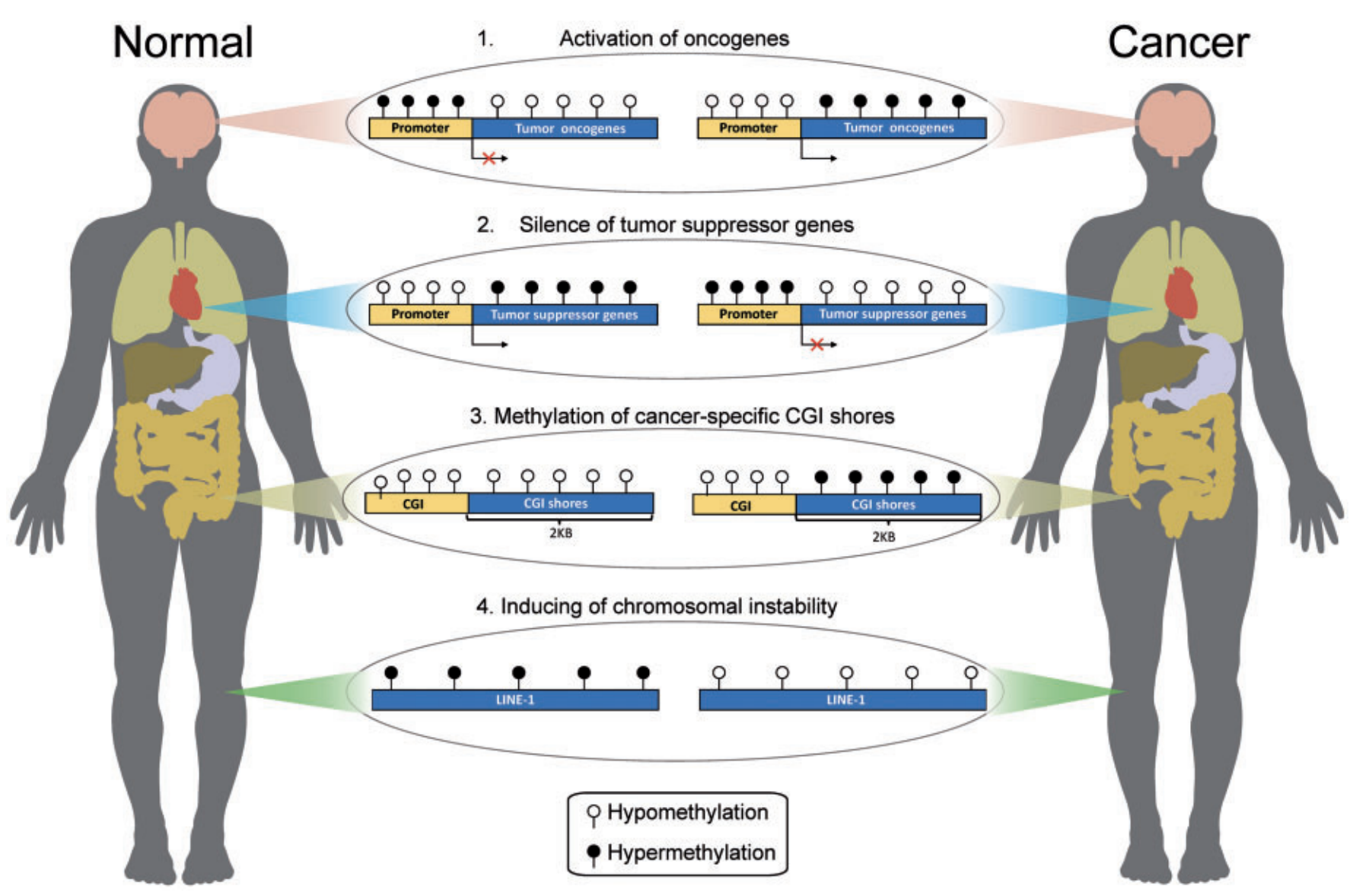 |
Epigenetic dynamics in mammalian development
| Epigenetic marking systems confer precise regulation of gene expression during mammalian development. DNA methylation and histone modification undergo dynamics during mammalian development (Liu et al. 2014 Database; Liu et al. 2013 Sci. Rep.). Using knockout mice of Dnmt3a/b, we demonstrated essential roles of DNA methylation in decommissioned fetal enhancers linking to kidney disease (Guan, Liu et al. 2020 JASN; Liu et al. In preparation). These studies highlighted locus-specific convergence of genetic, epigenetic, and developmental elements in disease development. | 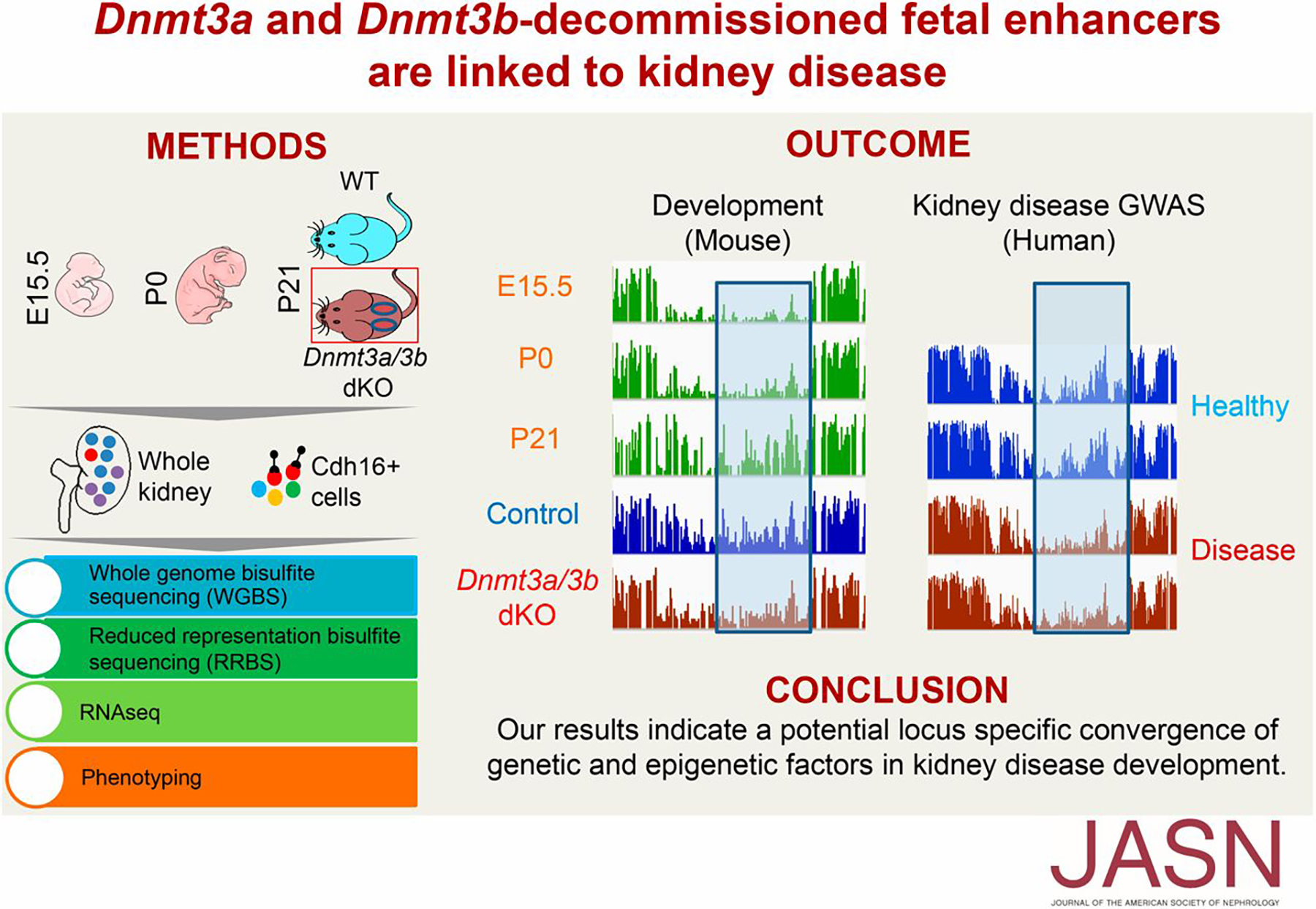 |
Cellular origins of age-related chronic diseases
| The causal cell type and regulatory mechanisms are poorly understood for complex diseases. To identify cell types causally associated with kidney disease, we generated single-nucleus transposase-accessible chromatin with sequencing (snATAC-seq), single-cell RNA sequencing (scRNA), and spatially resolved transcriptomics in mouse and human kidneys. The analysis of these single-cell resolution datasets illustrated the crucial roles of kidney cells (e.g., proximal tubule cells) and cell type-specific genes (e.g., SLC47A1) in kidney injury and fibrosis. (Liu et al., 2022 Nature Genetics; Sheng,…,Liu et al., 2021 Nature Genetics; Miao,…, Liu et al. 2021, Nature Comms; Doke,…, Liu et al., 2021 JCI; Dhillion,…, Liu et al., Cell Metabolism; Amin,…, Liu et al. 2022 BioRxiv). | 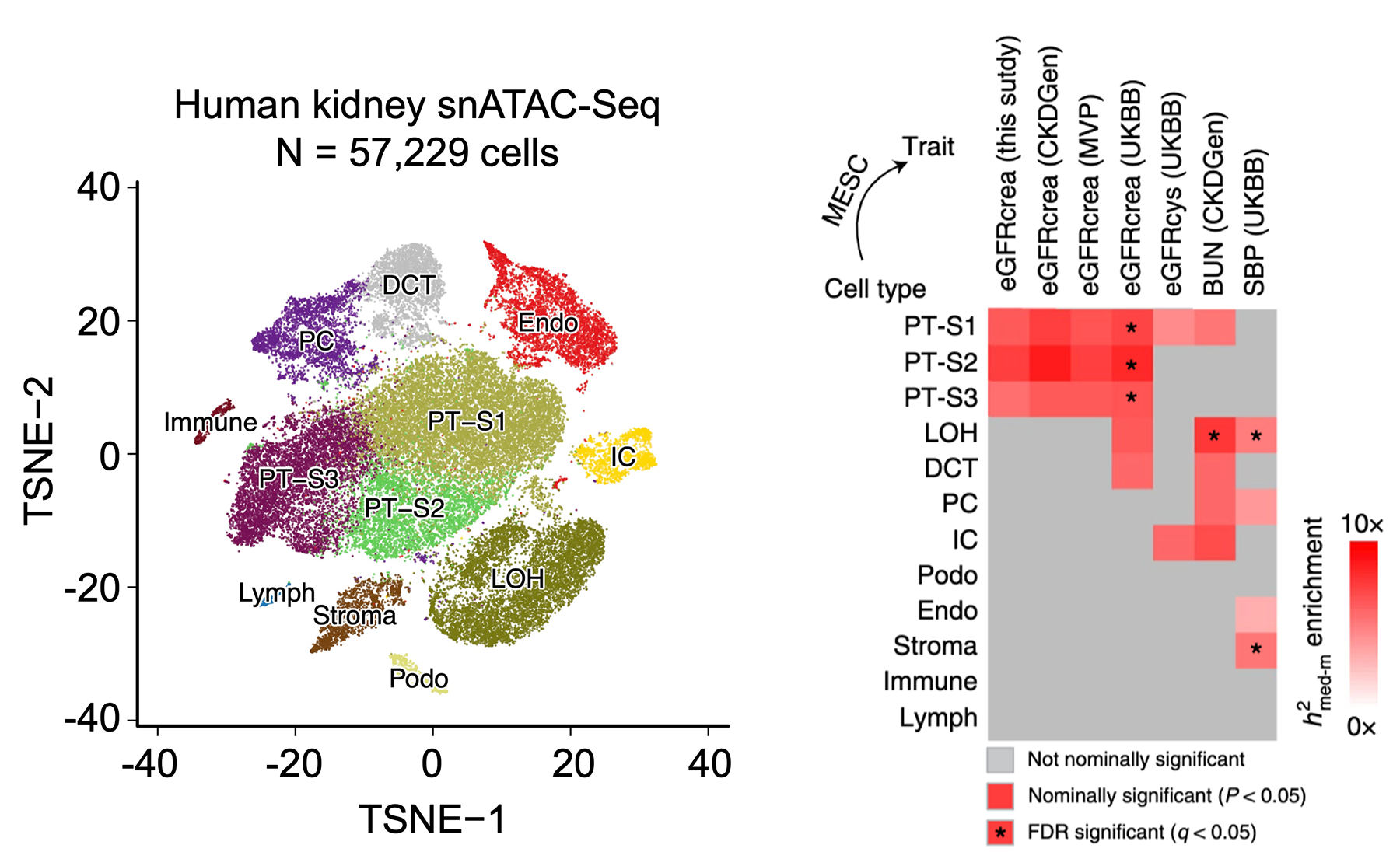 |